CRISPR method – easier and more precise genome engineering
 A group of researchers from MIT, the Broad Institute, and Rockefeller University, have developed a new technique for precise genetic alteration of living cells by inserting or deleting genes. In order to create new genome-editing technique, the researchers modified a set of bacterial proteins that normally protect bacteria from viruses. According to researchers, the technology offers an easy-to-use and less-expensive way to engineer various organisms.
A group of researchers from MIT, the Broad Institute, and Rockefeller University, have developed a new technique for precise genetic alteration of living cells by inserting or deleting genes. In order to create new genome-editing technique, the researchers modified a set of bacterial proteins that normally protect bacteria from viruses. According to researchers, the technology offers an easy-to-use and less-expensive way to engineer various organisms.
“Using this system, scientists can alter several genome sites simultaneously and can achieve much greater control over where new genes are inserted”, said Feng Zhang, an assistant professor of brain and cognitive sciences at MIT and leader of the research team. “Anything that requires engineering of an organism to put in new genes or to modify what’s in the genome will be able to benefit from this.”
A little bit of genome engineering history
The first genetically modified mice were created in the 1980s by inserting small pieces of DNA into an early-stage mouse embryo. Genetically modified mice are used extensively in research as research models for human diseases. Small pieces of DNA are randomly added into the mouse genome, thus making the researches unable to target the newly delivered genes to replace the desired genetic sequence.
This phenomenon forced various research groups to seek the solution which enables them to precisely edit the genome. Many DNA inserting methods used for creation of genetically modified organisms rely on the process of homologous recombination. Homologous recombination is a fundamental biological process in which nucleotide sequences are exchanged between two similar or identical DNA molecules. Homologous recombination method is used when one allele is replaced with an engineered construct without affecting any other locus in the genome. This method can be performed only if you know the DNA sequence of the gene you want to replace. It involves delivering of the DNA sequence which includes the gene of interest alongside the flanking DNA that is identical to the targeted locus. However, in most organisms, homologous recombination is a rare event, thus making the success rate of this method very low.
More recently, researchers revealed that efficiency of this process could be improved by adding enzymes known as nucleases that are capable of cutting DNA. Zinc finger nucleases (ZFNs) are artificial restriction enzymes designed to cut at specific DNA sequences. ZFNs consist out of a zinc finger DNA-binding domain and a DNA-cleavage domain. Although zinc fingers commonly deliver the nuclease to a specific location, they can’t recognize every possible DNA sequence. Aside limited usefulness of this enzyme, it is also complicated to design and assemble.
On the other hand, newer complexes such as Transcription Activator-Like Effector Nucleases (TALENs) are far more effective than ZFNs, however, they can be expensive and difficult to design.
CRISPR genome engineering technology and its potential application
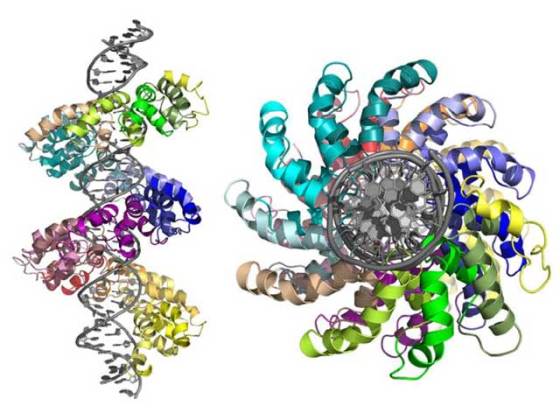
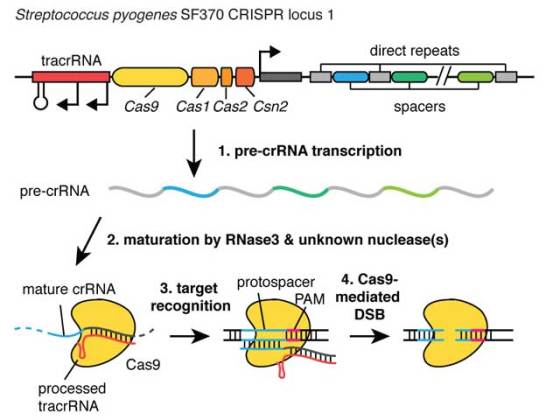
Inspired by naturally occurring bacterial protein-RNA systems that recognize and cut viral DNA, the researchers created the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) genome engineering method. CRISPR genome engineering relies on a bacterial nuclease called Cas9. Cas9 isbound to short bacterial RNA sequences which guide Cas9 nucleases to specific locations in the genome. Once they encounter a match, Cas9 cuts the DNA. This system can be used either to disrupt the gene function or to replace it with a new one.
This approach is far more precise and efficient in comparison to ZFNs and TALENs. Unlike aforementioned methods, Cas9 is not activated if there is a single base-pair mismatching between the RNA sequence and the genome sequence. Each of the RNA segments can target a different sequence, thus enabling nuclease to target one or more positions in the genome.
There is a broad range of potential applications for this new genome engineering technology. It could be used to design animal models to study human diseases, to engineer organisms that produce biofuels and to develop new therapies. Since it is more efficient, the technology can replace ZFNs in Huntington’s disease clinical trials.
The new technique might be used for HIV treatment by removing elements such as patient’s lymphocytes and mutating the CCR5 receptor which is used by the virus to enter its target cells. Once modified and returned back into the patient, these cells could resist HIV infection.
“Using this genome editing system, you can very systematically put in individual mutations and differentiate the stem cells into neurons or cardiomyocytes and see how the mutations alter the biology of the cells”, said Zhang.
MIT researchers tested the system in cells grown in the lab, but they plan to apply the new technology to study brain function and diseases. They created Genome Engineering Resources website with tools and advices for using this new method. The genetic components are widely available at the non-profit Addgene website, where researchers and other website members deposit the necessary genetic data.
Fore more information about the CRISPR genome engineering technology, read the article published in the journal Science: “Multiplex Genome Engineering Using CRISPR/Cas Systems” [1.63MB PDF].









Leave your response!