PaR-PaR – a biology-friendly robot programming language
 A collaboration between U.S. Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI) and Lawrence Berkeley National Laboratory (Berkeley Lab) resulted with a project named Programming a Robot (PaR-PaR) – a simple high-level, biology-friendly, robot-programming language that allows easier robot training. PaR-PaR could provide the researchers with ways to train the liquid-handling robots with greater ease and thereby enable experiments that otherwise might not have been considered.
A collaboration between U.S. Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI) and Lawrence Berkeley National Laboratory (Berkeley Lab) resulted with a project named Programming a Robot (PaR-PaR) – a simple high-level, biology-friendly, robot-programming language that allows easier robot training. PaR-PaR could provide the researchers with ways to train the liquid-handling robots with greater ease and thereby enable experiments that otherwise might not have been considered.
To date, automation companies have targeted the highly-repetitive industrial laboratory operations market while largely ignoring the development of flexible easy-to-use programming tools for dynamic non-repetitive research environments. As a consequence, researchers in the biological sciences have had to depend upon professional programmers or vendor-supplied graphical user interfaces with limited capabilities.
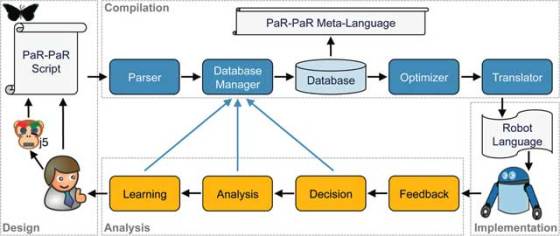
Led by Nathan Hillson, a biochemist at the U.S. DOE’s JBEI who directs JBEI’s Synthetic Biology program and also holds an appointment with the Berkeley Lab’s Physical Biosciences Division, the researchers developed PaR-PaR – a robot-programming language which uses an object-oriented approach that represents physical laboratory objects – including reagents, plastic consumables and laboratory devices – as virtual objects.
“The syntax and compiler for PaR-PaR are based on computer science principles and a deep understanding of biological workflows”, said Hillson. “After minimal training, a biologist should be able to independently write complicated protocols for a robot within an hour. With the adoption of PaR-PaR as a standard cross-platform language, hand-written or software-generated robotic protocols could easily be shared across laboratories.”
Hillson, who earlier led the development of j5 (a software for identifying cost-effective DNA construction strategies for a single robot platform), states that aside enabling biologists to manually instruct robots in a time-effective manner, PaR-PaR can be executed on many different robot platforms.
“Our vision was for a single protocol to be executable across different robotic platforms in different laboratories, just as a single computer software program is executable across multiple brands of computer hardware,” Hillson says. “We also wanted robotics to be accessible to biologists, not just to robot specialist programmers, and for a laboratory that has a particular brand of robot to benefit from a wide variety of software and protocols.”
Each object in a lab where a robot is used is described with associated properties, such as a name and a physical location, and multiple objects can be grouped together to create a new composite object with its own properties. Actions can be performed on objects and sequences of actions can be merged into procedures that in turn are issued as PaR-PaR commands. Collections of procedural definitions can be imported into PaR-PaR with use of external modules.
Using robots to perform labor-intensive multi-step biological tasks, such as the construction and cloning of DNA molecules, can increase research productivity and lower costs by reducing experimental error rates and providing more reliable and reproducible experimental data.
Hillson and his colleagues have developed PaR-PaR as open-source software which is freely available through its web interface on the public PaR-PaR webserver. The PaR-PaR software is also available at its github project page.
For more information, you can read the paper published in ACS Synthetic Biology: “PaR-PaR Laboratory Automation Platform” [1.3MB PDF].









Leave your response!